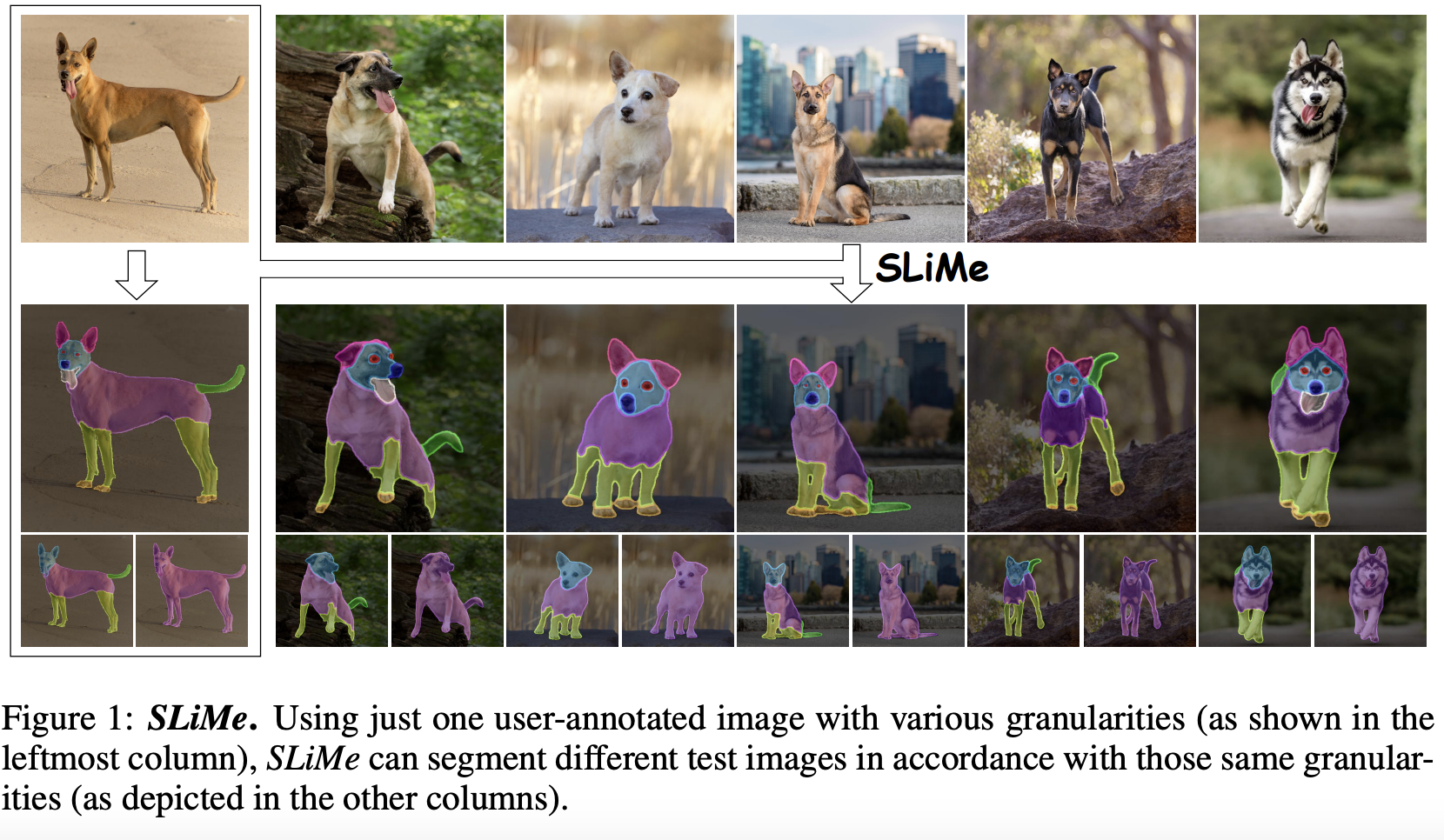
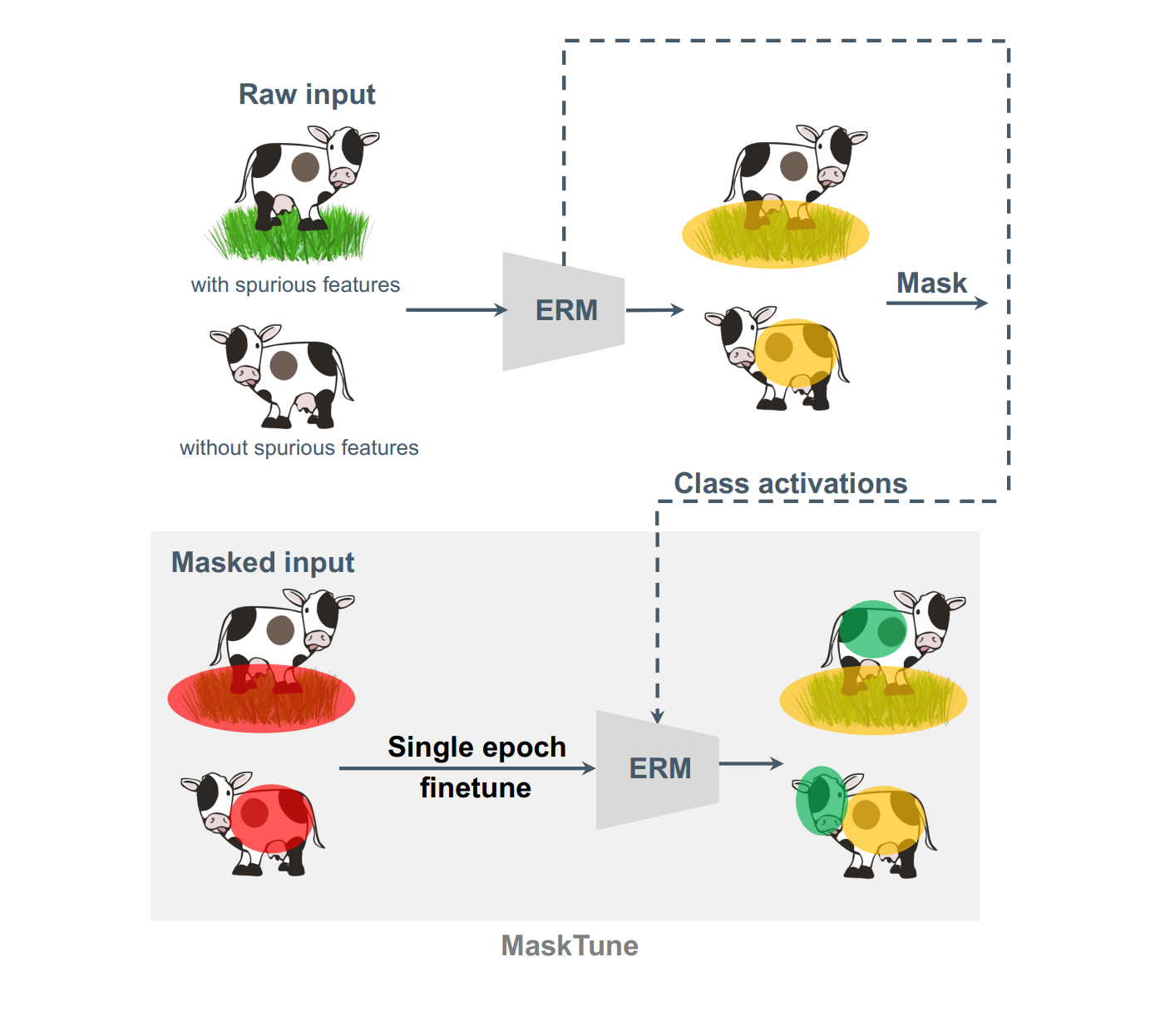
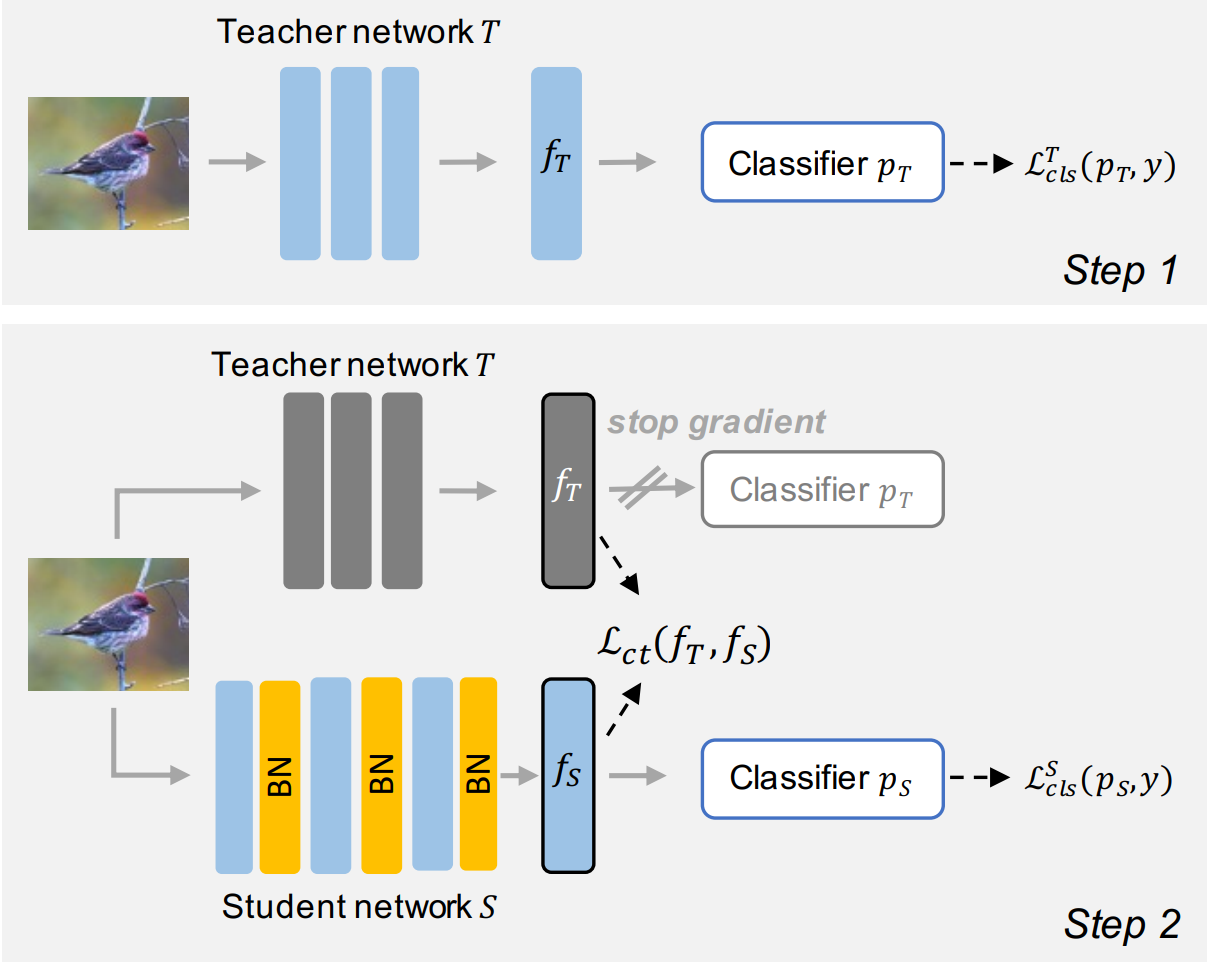
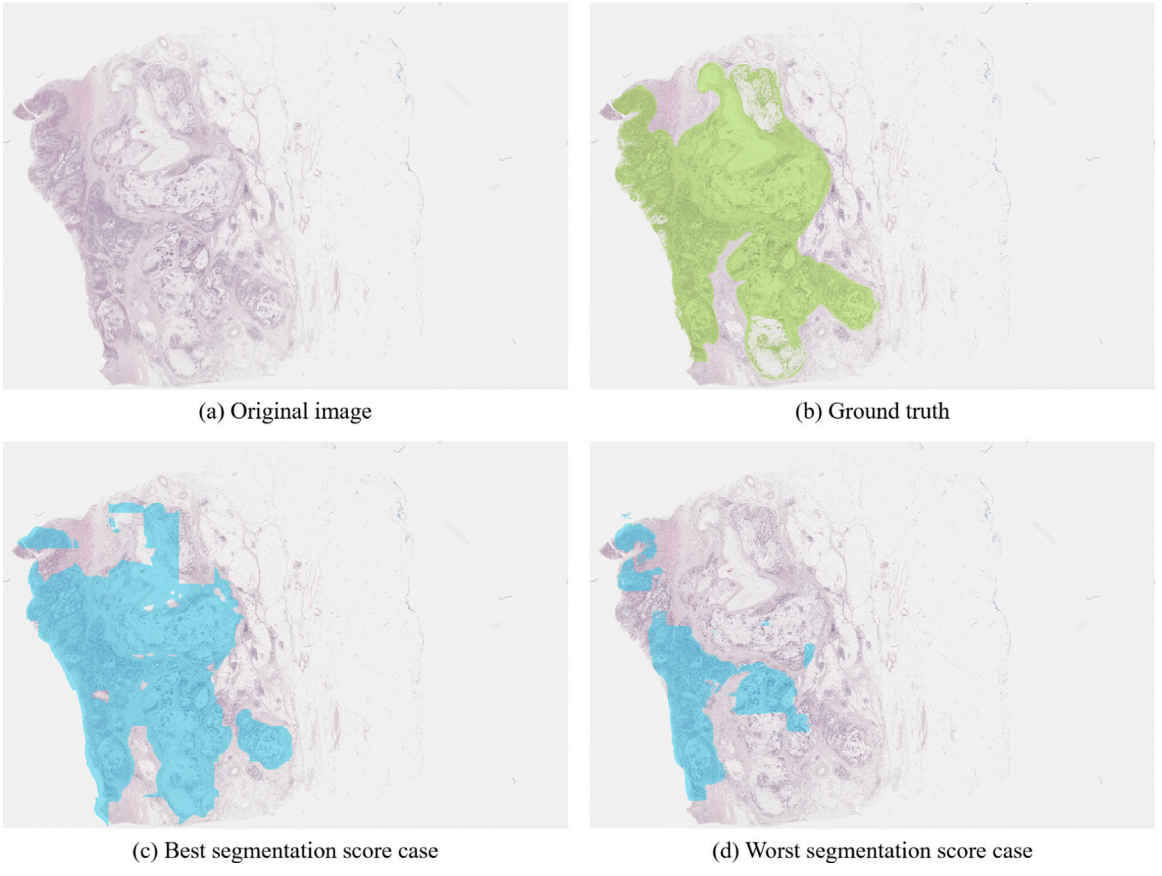
Aliasghar Khani
Machine Learning Researcher
Currently, I am pursuing a master's degree at Simon Fraser University, where I have the privilege of being supervised by Professor Ghassan Hamarneh, with Dr. Saeid Asgari as my co-supervisor. My academic journey began at Sharif University of Technology (SUT), where I completed my Bachelor's degree in Computer Engineering from 2016 to 2021, working under the guidance of Professor MohammadHossein Rohban for my B.S. project. During my B.S. project, I focused on leveraging self-supervised and weakly supervised methods to enhance adversarially trained models. This research aimed to address the challenges associated with adversarial training by exploring alternative learning approaches. Computer vision is my primary area of interest and research. I am particularly intrigued by the fundamental problems within this field, such as robustness and bias. Additionally, I have practical experience in combining language models with computer vision techniques, which has allowed me to explore the intersection of these domains. One of my greatest passions is continuous learning, and I am committed to utilizing my knowledge to tackle significant real-world problems, both through collaborative teamwork and individual efforts. In summary, my research interests encompass:
- Robustness
- Self supervised learning
- Generalization
My Skills
Coding skills
Python
C/C++
JavaScript
Matlab
Other Skills
Mathematics Knowledge
Machine Learning Knowledge
Machine Learning Frameworks
Education
-
2021 - Current
Computer Science
Simon Fraser UniversityM.Sc. student under supervision of Professor Ghassan Hamarneh
-
2016 - 2021
Computer Engineering
Sharif University of TechnologyB.Sc. in Computer Engineering
-
2012 - 2016
Mathematics and Physics
Shahid Madani High School
Experience
-
2022
AI Research Internship
AutoDeskUnder the supervision of Dr. Saeid Asgari
-
2020 - 2021
Research at Bioinformatics and Computational Biology (BCB) Lab
Sharif University of TechnologyUnder the supervision of Prof. Mohammad Hossein Rohban
-
2019 - 2021
AI Researcher
Iran's National Elites FoundationUnder the supervision of Prof. Mahdieh Soleymani Baghshah